Serum microRNAs (miRNAs) have recently emerged as promising biomarkers for a variety of diseases including cancer and metabolic disorders. Despite the increasing category of small RNAs, the existence and diagnostic value of other serum small RNAs are surprisingly few.
By analyzing small RNA deep-sequencing data (18-40nt) from animal sera, a research group led by Dr. Enkui Duan at Institute of Zoology, Chinese Academy of Sciences revealed that besides the previously well-characterized microRNA population, the serum also contains an ancient class of tRNA-derived small RNAs (tsRNAs) that abundantly, conservatively existed in a wide range of vertebrate species (from fish to human).
tsRNAs are stably existed in the serum, free from serum microvesicles (exosomes). The stabilization mechanism of serum tsRNAs in an RNase-rich blood environment involves at least two layers of protection: through binding and co-existed with serum protein complexes, as well as by nucleotide modifications inheriting from their tRNA predecessors.
Interestingly, serum tsRNAs showed a surge upregulation during LPS-induced acute inflammation in mouse and monkey, as well as in human patients under virus infection (HBV replication phase), suggesting their active involvements in infection-induced defensive response.
These data unveiled a hidden layer of serum small RNAs linking with disease condition, opening future avenues for the development of novel biomarker approaches based on analyzing serum tsRNAs.
The study entitled "Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection" was first published online in Journal of Molecular Cell Biology on Dec 30, 2013.
PDF Link 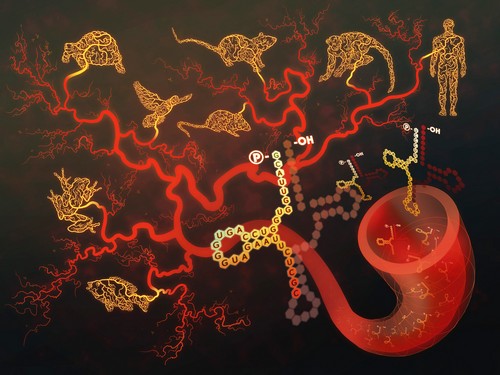
Discovery of an ancient class of tRNA-derived small RNAs (tsRNAs) abundantly and conservatively existed in the sera of a wide range of vertebrate species along the evolution tree. The serum tsRNAs showed sensitive response to active infection in mouse, monkey and human being.

