Hematopoietic stem cells (HSCs) are the only cells which have the capability to self-renew and differentiate into all blood lineages. The limit of the donors to provide sufficient HSCs in the clinic makes it urgent to produce HSCs ex vivo or in vitro in regenerative medicine. In vertebrate embryos, HSCs are derived from hemogenic endothelium (HE), a subset of endothelial cells (ECs) in the ventral wall of the dorsal aorta (DA), through a process named endothelial-to-hematopoietic transition (EHT). Then they expand in the caudal hematopoietic tissue (CHT, in zebrafish) or fetal liver (FL, in mammals), and finally home to the kidney (in zebrafish) or the bone marrow (in mammals) to sustain adult life. After decades of studies in this field, by using animal models from zebrafish, frog, chicken to mouse, scientists have achieved some basic understanding about developmental hematopoiesis in vivo and have tried to recapitulate it in vitro to some extent. However, the detailed regulatory mechanism underlying this cell fate change during EHT is still largely unclear, especially for the role of epigenetic modification in this process. N6-methyl-adenosine (m6A) is one of the most prevalent mRNA modifications in eukaryotes. Our knowledge on its functions is based on the identification and characterization of m6A "writers" (methyltransferase complex including METTL3, WTAP and METTL14), "readers" (RNA binding proteins like YTH family members) and "erasers" (demethylase including FTO and ALKBH5). Recently, a series of seminal studies have begun to reveal its important functions including the maternal-to-zygotic transition in zebrafish, sex determination in drosophila and T cell homeostasis in mouse. However, the physiological functions and regulatory mechanism of m6A modification, especially during embryogenesis in vertebrates still remain elusive. The Hematopoietic and Cardiovascular Development Group led by Prof. Feng Liu utilizes two model systems including zebrafish and mouse to study the regulatory mechanism of HSC development. In collaboration with Prof. Yungui Yang from Beijing Institute of Genomics, they previously identified the m6A methyltransferase complex in zebrafish (Cell Res, 2014). Based on these findings, these two collaborative groups performed m6A-Seq in zebrafish embryos, and found that the m6A enrichment on embryonic development-related mRNAs was significantly decreased upon the loss of mettl3. Moreover, mettl3 is specifically expressed in the hemato-vascular systems in zebrafish embryos, indicating a potential function of METTL3-mediated m6A modification in hematopoietic development. Further detailed studies revealed that, upon mettl3 depletion, the endothelial cell fate was enhanced, but the EHT was blocked, thereby leading to the failure of HSC generation. Bioinformatics analysis of m6A-Seq and RNA-Seq revealed that the m6A modification on a series of arterial endothelial cell related genes, especially notch1a mRNA, was decreased, while their mRNA levels were increased. Furthermore, the YTHDF2-RIP-Seq and single-nucleotide resolution m6A-miCLIP-Seq demonstrated that YTHDF2 mediates m6A-modified notch1a mRNA stability, supporting that m6A functions as a rheostat to maintain the balance of gene expression between endothelial cell and HSC during EHT. These findings were also confirmed in mouse experiments, suggesting the evolutionally conserved function of m6A in HSC development across vertebrates. This study, for the first time, uncovers the regulatory mechanism of m6A mRNA methylation in vertebrate hematopoietic stem cell fate determination, and also provides useful insights to the in vitro generation of HSCs for clinical application. This paper entitled "m6A modulates haematopoietic stem and progenitor cell specification" was published online in Nature on Sep 6, 2017 (Nature, doi: 10.1038/nature23883). This work was supported by grants from the National Natural Science Foundation of China, and the Ministry of Science and Technology of China and the Strategic Priority Research Program of the Chinese Academy of Sciences. http://www.nature.com/nature/journal/vaop/ncurrent/full/nature23883.html 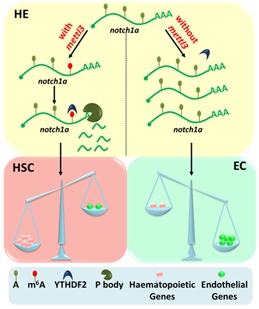
Figure 1. Schematic representation of the role of m6A methylation in HSC fate determination during EHT via METTL3-notch1a. 
Figure 2. The artistic image shows hematopoietic stem cell (blue) budding from hemogenic endothelial cell (green) through the endothelial-to-hematopoietic transition (EHT), which is modulated by m6A modification (pink) of Notch1a mRNA (brown) to ensure hematopoietic stem cell specification. Contact: Feng Liu, PhD State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Chaoyang District, Beijing 100101, P.R.China Email: liuf@ioz.ac.cn
|


