DNA methylation is one of the major epigenetic modifications in vertebrates, which involves adding a methyl group to the 5th carbon of cytosine to form 5-methylcytosine (5mC), which is established and maintained by DNA methyltransferases (DNMT). DNA methylation is associated with a number of key processes including transcription regulation, chromosome stability, genomic imprinting, and X-chromosome inactivation. Genome-wide 5mC mapping has implicated the role of DNA methylation during early embryogenesis. However, the precise role of DNA methylation in vertebrate development is still not fully understood.
Establishment of the three body axes (anterior-posterior, dorsal-ventral and left-right axes) is a critical step during animal development. Vertebrate body display external symmetry but the internal organs are positioned asymmetrically such as the heart, pancreas, liver and intestines having a defined position within the body cavity. Left-Right asymmetry is first established by breaking symmetry, and then followed by laterality organ formation, where cilia generate directional fluid flow, which leads to asymmetric gene expression. This asymmetric expression pattern then will direct internal organ primordium to position asymmetrically. However, very little is known about the role of DNA methylation in LR determination.
Using zebrafish as a model, the Hematopoiesis and Cardiovascular Development Group led by Prof. Feng Liu, found that loss of dnmt1 or dnmt3bb.1 disrupts laterality of organs including heart, pancreas, and liver. Mechanistically, hypomethylation of the lefty2 gene enhancer caused by loss-of-dnmt1 can promote lefty2 expression, which in turn inhibits Nodal signaling, therefore leading to impaired DFC specification and loss of LR asymmetry. In addition, Dnmt3bb.1 is required for cadherin 1 (cdh1)-mediated DFC clustering to ensure proper LR determination. This study unravels a new layer of regulation involving dynamic DNA methylation in embryonic development in vertebrates.
This study ‘Epigenetic regulation of left-right asymmetry by DNA methylation’ was published in EMBO J online on Sep 7, 2017. This work was supported by grants from the National Natural Science Foundation of China and the Ministry of Science and Technology of China.
Website:http://emboj.embopress.org/content/early/2017/09/07/embj.201796580
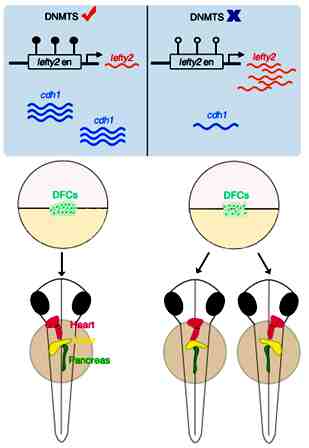
Model for regulation of LR asymmetry by DNA methylation during embryogenesis in vertebrates
In the presence of DNMTs, the expression of lefty2 and cdh1 is tightly controlled to ensure normal LR asymmetry; in the absence of DNMTs, the expression of lefty2 and cdh1 is dysregulated due to hypomethylation, thereby causing abnormal LR asymmetry.

