The harsh environment of the Qinghai-Tibet Plateau, the highest and largest plateau in the world, exerts severe selective pressure that has resulted in the evolution of similar phenotypic adaptations in humans, mammals, and birds (e.g. hypoxic resistance, cold tolerance, enhanced metabolic capacity and increased body masses). However, genomic studies have shown that different organisms adapt to high altitudes via multiple genetic routes, which may be largely subject to phylogenetic context. Comparisons of phylogenetically distinct species in previous studies may have underestimated the actual degree of similarity in genomic evolution. In addition, although high-altitude adaptation has been well investigated at the gene sequence level, studies of gene expression across multiple tissues across multiple species are rarely concerned.
Recently, a study entitled “Comparative transcriptomics of 3 high-altitude passerine birds and their low-altitude relatives” was published in PNAS on May 20, 2019. The research team led by Prof. LEI Fumin from Institute of Zoology of the Chinese Academy of Sciences, compared transcriptomic data of three high-altitude passerine birds with those of their low-altitude relatives by integrating protein-coding sequence comparison and gene expression analysis across multiple tissues to explore high-altitude adaptation within a phylogenetic framework.
Sequence comparison revealed that the three high-altitude birds were genetically convergent on positively selected genes, but their amino acid substitutions mostly occurred at divergent sites, suggesting that adaptive convergence seldom occurred at the amino acid substitution level. Gene expression analysis found that the high-altitude environment might have promoted substantial expression shifts in the three high-altitude birds since expression profiles of differentially expressed genes and altitude-associated genes (altitude-clustered pattern) largely differed from those of all genes (tissue-clustered pattern). Few genes under positive selection for all three high-altitude species overlapped with differentially expressed genes, but the interaction between gene expression and altitude and the interaction between gene connectivity and altitude were correlated with evolutionary rates of genes.
Taken together, these new findings suggested that the high-altitude birds might evolve in a concerted way through both sequence changes and expression shifts.
This study was funded by grants from the Strategic Priority Research Program, Chinese Academy of Sciences and the National Science Foundation of China.
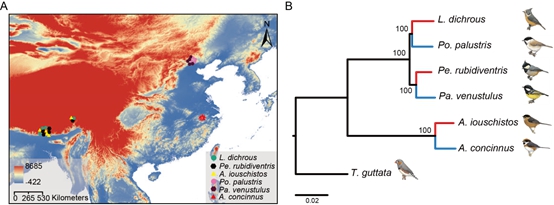
Figure 1. (A) Sampling locations. (B) Maximum likelihood phylogeny of the six studied species with the zebra finch (Taeniopygia guttata) as an out-group. Red branches indicate the high-altitude species; blue branches indicate the low-altitude species. (Image by IOZ)
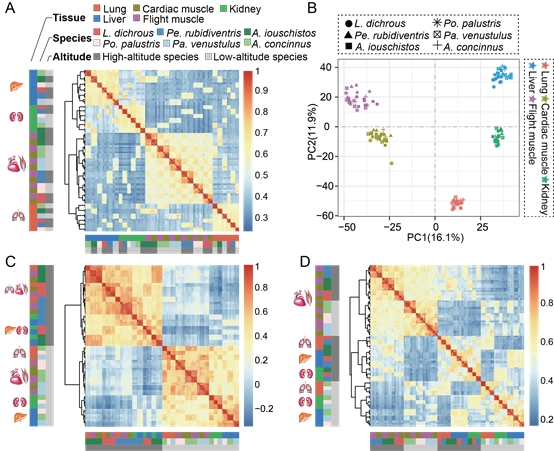
Figure 2. Gene expression patterns across the six species. (A) Symmetrical heat map of Spearman's correlation coefficients between all pairs of samples across all genes. (B) PCA of the log-transformed normalized expression levels of all orthologs across all species and tissues. (C) Symmetrical heat map of Spearman's correlation coefficients between all pairs of samples across differentially expressed genes. (D) Symmetrical heat map of Spearman's correlation coefficients between all pairs of samples across altitude-associated genes. (Image by IOZ)


