Chromosome evolution is an important driver of speciation and species evolution. Carnivora species exhibit sharply contrasting karyotypes and provide excellent examples for studying chromosome evolution. Although previous studies have established a series of comparative chromosome maps in different groups of Carnivora species using chromosome painting method, these cytogenetic methodologies do not provide sufficient resolution to perform genome-scale synteny analysis or to accurately identify homologous synteny blocks (HSBs), fine-scale rearrangements, and evolutionary breakpoint regions (EBRs).
By combining the strategies of 10X Genomics Chromium, flow-sorting and reference-assisted assembly, recent study presented a chromosome-level giant panda draft genome. Based on this giant panda (2n=42), published dog (2n=78) and cat (2n=38) chromosome-level genomes, the researchers conducted six large-scale pairwise synteny alignments and identified a total of 59, 37 and 55 EBRs in giant panda, dog and cat genomes. Significant increases for gene density, GC content and repetitive content were observed in giant panda and dog EBRs, as compared with the whole genomes.
The gene functional enrichment analysis showed that for all of three Carnivora genomes, some genes located in EBRs were significantly enriched in pathways or terms related to sensory perception of smell. In addition, the intact sweet receptor gene TAS1R2, which located in the EBR of giant panda genome, has become a pseudogene in the cat genome, suggesting that chromosome rearrangement may play a role in the TAS1R2 pseudogenization in cat genome. These findings show that olfaction evolution may be affected by chromosome rearrangement events.
This research has been published on December 6, 2019 as a research paper entitled “Chromosome-level genome assembly for giant panda provides novel insights into Carnivora chromosome evolution” in Genome Biology. Dr. Huizhong Fan and Prof. Qi Wu are the co-first authors, and Prof. Yibo Hu is the corresponding author. Prof. Fuwen Wei provided a lot of guidance and help for this work. The research was funded by the National Natural Science Foundation of China, the Chinese Academy of Sciences, and etc.
Article link:https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1889-7
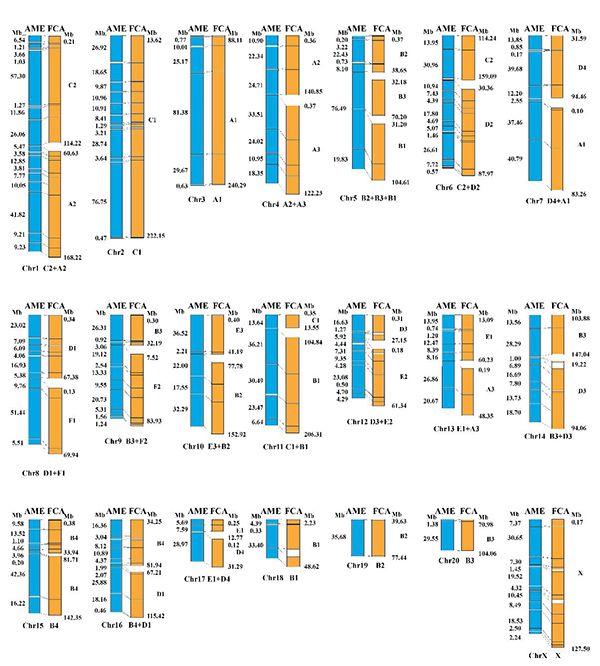
Fig.1 The synteny alignment of giant panda (AME) and cat (FCA) chromosome-level genomes
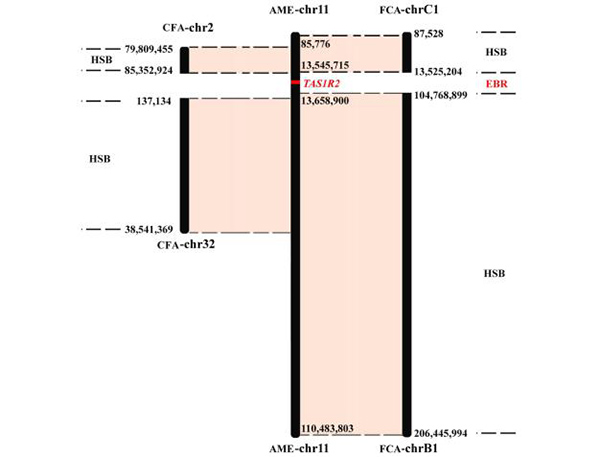
Fig.2 A case of evolutionary breakpoint region related with the sweet receptor gene TAS1R2


