Classical quantitative genetics has found that most of phenotypes are polygenic traits. Under this polygenic model, natural selection often acts on many loci simultaneously, resulting in the combination of a few loci with major effects and many loci with small effects controlling adaptive changes in phenotypes, which presents challenges to our understanding of genetic basis underlying polygenic traits. Avian beak is a typical polygenic trait, which is the consequence of the variety of functions that beaks serve (e.g. foraging, preening, nest-building, cavity excavation) and the diversity of habitats where birds live. As a result, genetic basis of beak morphology was well studied in chickens and Darwin’s finches, yet remains poorly understood in wild birds. Due to the characteristics of being more likely to be fixed by strong selective coefficients and less susceptible to loss by genetic drift, major loci may be more relevant to phenotypes and easier to be detected from the genome.
Prof. Fumin Lei’s team from the Institute of Zoology, Chinese Academy of Sciences conducted phylogenomes of 14 tits species (Paridae) and found a major locus (COL27A1) that effects the beak elongation of the Ground Tit (Pseudopodoces humilis) as response to ground-foraging and cavity-nesting habits on the Qinghai-Tibet Plateau (Figure 1). This study entitled “Comparative genomics reveals evolution of a beak morphology locus in a high-altitude songbird” was published online in Molecular Biology and Evolution on June 27th, 2020.
The study combined two methods, i.e., fixation index (FST) and partial mantel test (PMT), to perform genome-wide analyses across 14 parid species, and identified 25 highly divergent genomic regions that are significantly associated with beak length, finding seven candidate genes involved in bone morphogenesis and remolding (Figure 2). Neutrality tests (nucleotide diversity, Tajima’s D、Fu & Li’s D and Zeng’s E) indicated that a model allowing for a selective sweep in the highly conserved COL27A1 gene best explains variation in beak length. We also identified two non-synonymous fixed mutations in the collagen domain that are predicted to be functionally deleterious yet may have facilitated beak elongation (Figure 2). Although this study did not do further functional experimental validation, it provides important insights into understanding the genetic mechanism of beak elongation and its role in the high-altitude adaptation, as well as highlights the importance of comparative genomics to studies of ecological adaptation and evo-devo.
Yalin Cheng, Matthew J Miller, Dezhi Zhang, Gang Song, Chenxi Jia, Yanhua Qu, Fumin Lei*. Comparative genomics reveals evolution of a beak morphology locus in a high-altitude songbird. Molecular Biology and Evolution, 2020. https://academic.oup.com/mbe/article/doi/10.1093/molbev/msaa157/5864032.
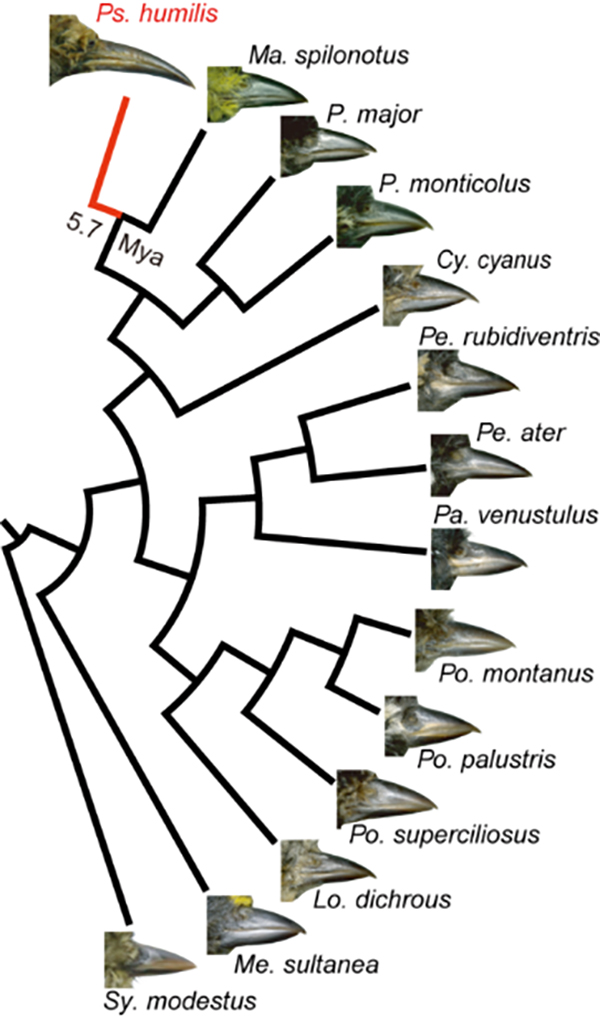
Figure 1. Comparison of beak morphology across 14 parid species
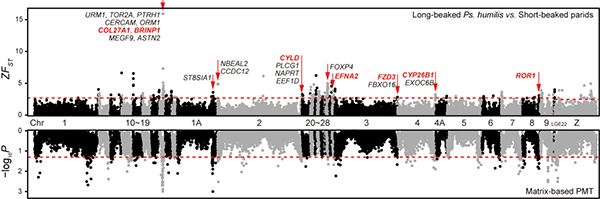
Figure 2. Candidate genes indentified by FST and PMT analyses.
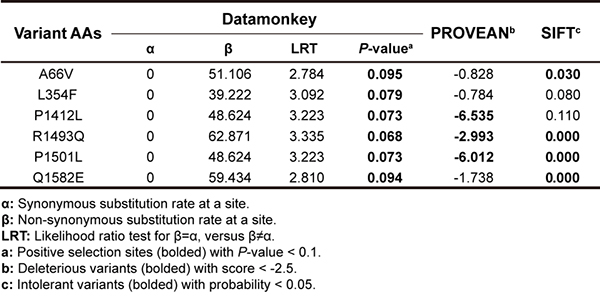
Figure 3. Selection analysis and function prediction for fixed non-synonymous sites.



