During early embryonic development, epigenetic landscapes undergo dramatic changes, which regulate cell fate transitions into different lineages. Human embryonic stem cells (hESCs) can be differentiated into multiple cell types, providing a highly tractable model system for studying mechanisms of early human development. Moreover, direct reprogramming of somatic cells into induced pluripotent stem cells (iPSCs) requires the erasure of original epigenetic modifications across the genome and the reestablishment of new pluripotency-related epigenetic states. DNA-RNA hybrids (R-loops) play key roles in both physiological and diseased states by regulating chromatin and genome organization. However, their homeostasis in cell differentiation and cell plasticity remains elusive. Therefore, a comprehensive and interactive atlas profiling R-loops in the context of key epigenetic marks, such as the transcriptome, histone modifications, DNA modification, and chromatin accessibility, in a robust and tractable human cellular model system may provide systematic information to reveal R-loop changes in association with other epigenetic modifications. Recently, scientists from the Institute of Zoology of the Chinese Academy of Sciences (CAS), Tsinghua University and Beijing Institute of Genomics of CAS have worked jointly and systematically characterized R-loops, DNA methylation, histone modifications, and chromatin accessibility in human pluripotent cells and their differentiated derivatives. Notably, they demonstrated a multi-faceted role of R-loops in cell fate determination that may serve as an additional layer of modulation on cell fate memory and cell plasticity. This study entitled "Genome-wide R-loop landscapes during cell differentiation and reprogramming" has been published online in Cell Reports on July 7th, 2020. To create an isogenic human stem cell platform, the researchers analyzed hESCs and four hESC-derived lineages (neural, mesenchymal, endothelial, and smooth muscle cells), as well as hiPSCs reprogrammed from hESC-derived hNSCs in parallel. Specifically, they profiled R-loops in a genome-wide strand-specific manner, together with seven reference profiles of major histone modifications (H3K27ac, H3K4me3, H3K36me3, and H3K27me3), transcriptome, DNA methylation, and chromatin accessibility, during in vitro lineage specification and cell reprogramming. They found that hESC-specific R-loops formed co-transcriptionally at pluripotency-related genes including OCT4 and NANOG. In differentiated cells, a group of R-loops were associated with increased repressive chromatin mark, H3K27me3 at pluripotency-related genes and other undesired lineage-restrictive genes, by which R-loops may participate in the inactivation of their expression. In addition, in hESC-derived lineages, newly formed R-loops occurred with active markers at lineage-controlling genes, thus likely promoting differentiation. During reprogramming, R-loops were only partially reset to an hESC-like state with almost one-third of R-loops persisting as “epigenetic memory” at early passage iPSCs. Further studies revealed that such residual R-loops could be erased by serial passaging, which was potentially related to further epigenetic remodeling of the cells. Altogether, researchers systematically investigated genome-wide R-loop landscapes and corresponding multi-layer transcriptional and epigenetic reference profiles during lineage specification and cell reprogramming. They found that R-loops were widely involved in cell fate determination and might serve as a novel form of epigenetic memory. These findings suggest that the elimination of unscheduled R-loops may be critical to the improvement of iPSC reprogramming quality, providing new insights into iPSC-based regenerative medicine. 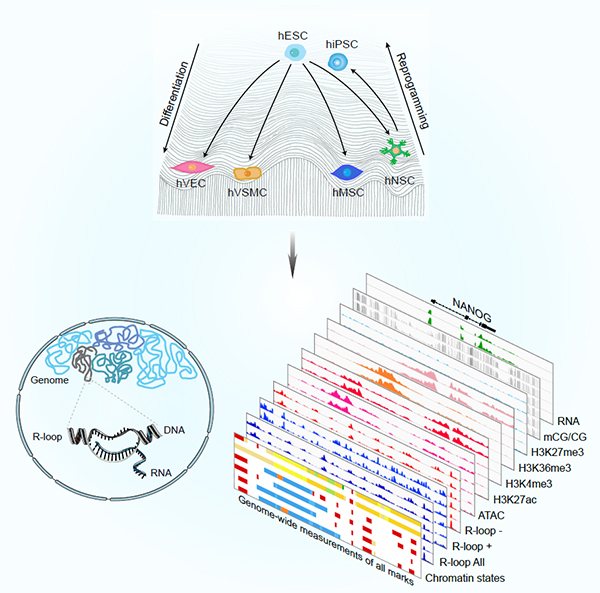
Figure. Genome-wide R-loop landscapes during cell differentiation and reprogramming
|

