X chromosome inactivation (XCI) is an important epigenetic event during early mammalian development. One of the two X chromosomes is randomly inactivated to ensure that gene expression level is consistent between XX and XY individuals. The X chromosome status in human early embryos is dramatically different from mouse. In addition, neither primed nor naive human embryonic stem cells (hESCs) were able to fully recapitulate X chromosome status in human preimplantation epiblast and they failed to execute random X chromosome inactivation (XCI) upon differentiation. Therefore, the study of human XCI has been hampered by the lack of proper models.
On July 15th, 2020, Haoyi Wang’s team from the Institute for Stem Cell and Regeneration, State Key laboratory of Stem Cell and Reproductive Biology, Institute of Zoology, Chinese Academy of Sciences, published a paper entitled” Overcoming autocrine FGF signaling-induced heterogeneity in naive human ESCs enables modeling of random X chromosome inactivation.” This study showed for the first time that human random XCI can be adequately modeled using homogenous naive hESCs resembling human preimplantation epiblasts (Figure 1).
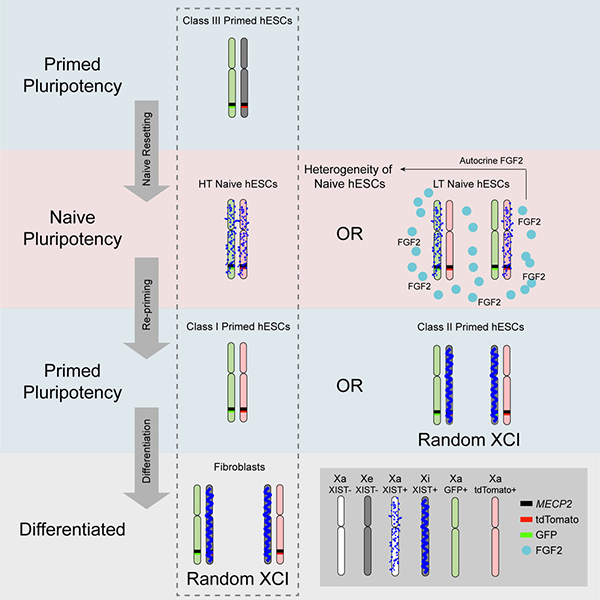
Figure 1. Modeling human random XCI using naive hESCs.
The study showed that within naive hESC culture, two populations of cells (HT, highly expressing TFCP2L1; LT, lowly expressing TFCP2L1) residing in different pluripotent states. Both HT and LT naive hESCs had two active X chromosomes, while exhibited different XIST expression patterns. HT naive hESCs showed bi-allelic XIST expression that is similar with human preimplantation epiblasts, while LT naive hESCs expressed XIST mono-allelically. The transcriptome of HT naive hESCs resembled that of human preimplantation epiblast to a greater extent than that of LT cells. Further exploring the underlying mechanism, they uncovered that incomplete blocking of autocrine FGF2 released by LT cells led to this heterogeneity.
In order to model human XCI, the authors re-adapted the HT naive hESCs into the primed culture medium (re-priming) and random XCI was initiated in a portion of cells, leading to the derivation of both pre- and post-XCI primed hESCs. Further in vitro differentiation of pre-XCI primed hESCs resulted in complete random XCI.
Taken together, through carefully delineating the heterogeneity of naive hESCs and testing different differentiation methods, Wang and his colleagues derived homogenous naive hESCs and established a robust platform for studying the process and mechanism of human XCI. This platform will be useful for the identification of key genes and pathways involved in X chromosome regulation in preimplantation epiblast, XCI initiation upon implantation, X chromosome choice during XCI, as well as XCI execution and maintenance in human.
Chenrui An, a Ph.D. student and associate professor Guihai Feng of the Institute of Zoology, Chinese Academy of Sciences are co-first authors of this paper. Professor Haoyi Wang from The State Key Laboratory of Stem cell and Reproductive Biology, Institute of Zoology, Institute for Stem Cell and Regeneration, Chinese Academy of Sciences is the corresponding author. At the same time, this research also benefited from the vigorous collaboration of Academician Qi Zhou from The State Key Laboratory of Stem cell and Reproductive Biology, Institute of Zoology, Institute for Stem Cell and Regeneration, Chinese Academy of Sciences, and Professor Falong Lu from the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences.

