Embryogenesis in animals is initially directed by maternal factors, including maternal mRNAs and proteins, deposited in the egg [1]. During the maternal-to-zygotic transition (MZT) process, the maternal transcripts undergo orchestrated translation and well-controlled clearance, and the animal begins to synthesize zygotic factors depending on embryonic genome. Afterwards, the zygotic factors replace the maternal factors and regulate subsequent individual development [1-3]. Previous study by Yang et al showed that RNA modification of m5C regulates maternal RNA stability [4]. However, the global mechanism for the regulatory program of maternal transcriptome remains largely unknown.
Recently, researchers from the Beijing Institute of Genomics of the Chinese Academy of Sciences and Tsinghua University and Institute of Zoology have collaborated to uncover the role of RNA structural dynamics in MZT and early embryogenesis regulation. This study entitled “RNA structural dynamics regulate early embryogenesis through controlling transcriptome fate and function” was published online in Genome Biology.

RNA structure is tightly involved in a myriad of posttranscriptional regulation on a global scale. In this study, researchers establish the global map of four nucleotide-based RNA structures during zebrafish early embryogenesis by icSHAPE (in vivo click selective 2’-hydroxyl acylation and profiling experiment) [5]. Strikingly, they observe that RNA structurally variable regions are enriched in the 3′ UTR and contain cis-regulatory elements important for MZT. Furthermore, they find that the RNA binding protein Elavl1a stabilizes some maternal mRNAs by binding to the cis-elements. Conversely, RNA structure formation suppresses Elavl1a’s binding leading to the decay of its maternal targets. These findings uncover that Elavl1a regulates maternal RNA stability in an RNA structure-dependent fashion.
Overall, this study reveals a broad and fundamental role of RNA structure-based regulation in vertebrate early embryogenesis.
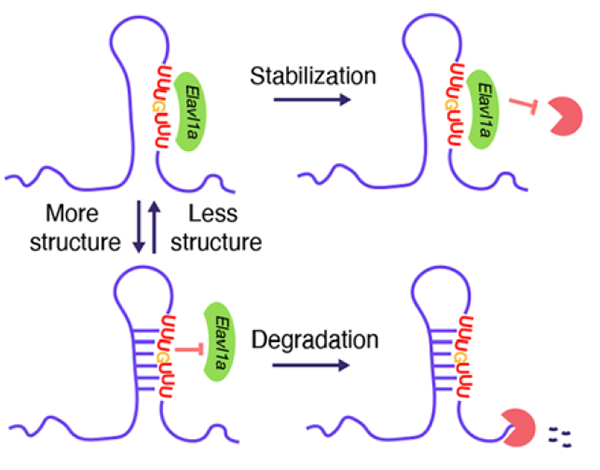
Fig.1 Schematic model shows that Elavl1a regulates RNA stability in a structure-dependent fashion
The link for this article: https://doi.org/10.1186/s13059-020-02022-2
References:
1.Lee MT, Bonneau AR, Giraldez AJ: Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 2014, 30:581-613.
2.Tadros W, Lipshitz HD: The maternal-to-zygotic transition: a play in two acts. Development 2009, 136:3033-3042.
3.Abrams EW, Mullins MC: Early zebrafish development: it's in the maternal genes. Curr Opin Genet Dev 2009, 19:396-403.
4.Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, Sun BF, Li A, Xia J, Chen J, et al: RNA 5-Methylcytosine Facilitates the Maternal-to-Zygotic Transition by Preventing Maternal mRNA Decay. Mol Cell 2019.
5.Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, Chang HY: Structural imprints in vivo decode RNA regulatory mechanisms. Nature 2015, 519:486-490.
(Contact: Feng Liu, liuf@ioz.ac.cn)


