Two or more species affect each other's evolution through the process of natural selection, which is called coevolution. It includes mutually beneficial coevolution and antagonistic coevolution, among which coevolution between host and parasite is typically antagonistic. For the previous studies on the genetic basis of coevolution, most of them focused on the candidate gene level and lacked genome-level study. With the development of next-generation genome sequencing technology, the genomes of many parasites that are closely associated with the health of human and domestic animals have been sequenced. However, most of them only focused on the structure and function of parasite genomes, but did not explore host genome-related changes and the coevolution mechanisms between host and parasite.
In order to explore the above issues, academician Fuwen Wei’s research team investigated the genomic mechanisms of coevolution between non-model mammals and their parasitic roundworms, adaptation mechanism of parasitic roundworms and special adaptation mechanism of panda roundworms through the de novo genome sequencing and assembly of giant panda roundworm (Baylisascaris schroederi), red panda roundworm (Baylisascaris ailuri), and lion roundworm (Toxascaris leonina).
The genome-wide species phylogenetic trees revealed that these parasitic roundworms have not phylogenetically coevolved with their hosts. After constructing genome-wide protein interactions between host and roundworm through simulating the life cycle of the roundworm within the host, we found that the immunoregulatory proteins CTSZ and P4HB played a central role in protein interaction between mammals and their parasitic roundworms. Furthermore, phylogenetic analyses of 186 pairs of interactive proteins for mammals and parasitic roundworms showed that seven pairs of interactive proteins had consistent phylogenetic topology, i.e., CRP-PLA2G1B, PLA2G7-PLA2G1B, PLA2R1-PLA2G1B, PLB1-PLA2G1B, IGFBP7-P4HB, MMP8-CTSZ and QPCT-CTSZ, suggesting the gene-level coevolution between hosts and parasites. These coevolutionary proteins were particularly relevant to immune response during the parasite infection.
Besides, to identify the specific amino acid substitutions of parasitic roundworms. the sequences of positively selected genes of parasitic roundworms were compared among 75 parasitic nematodes and 12 free-living nematodes. A total of 34 positively selected genes with 60 parasite-specific amino acid substitutions were identified, which were partly enriched in the larva development and reproduction-related pathways. Finally, we found that the roundworms of both pandas exhibited higher proportions of metallopeptidases, and more genes of glycosyltransferase and cytochrome P450 pathways occurred in their secretomes, which may be related to their adaptation to secondary metabolites of the host’s bamboo diet. The positively selected genes of the two panda roundworms were partly enriched in the pathways of larva development and reproduction, which may be related to fast larvae development of panda roundworms.
Our findings reveal the genetic basis of the coevolution between non-model mammals and their parasitic roundworms, and provide insights into the genetic mechanisms of coevolution between mammals and parasites. Additionally, the roundworm genome resources, secretomes and key interactive proteins of coevolution from this study offer key candidate gene resources and scientific basis for ascariasis prevention in both pandas.
The findings have been published online entitled "Genomic signatures of coevolution between non-model mammals and parasitic roundworms" in Molecular Biology and Evolution. Prof. Yibo Hu and Ph.D. student Lijun Yu are the first co-authors, and academician Fuwen Wei is the corresponding author. This research has been supported by the Strategic Priority Research Program of Chinese Academy of Sciences (CAS), the National Natural Science Foundation of China (NSFC), the Frontier Key project of CAS, and the Youth Innovation Promotion Association, CAS.
Paper link:
https://academic.oup.com/mbe/advance-article/doi/10.1093/molbev/msaa243/5910001?guestAccessKey=49baeb83-4656-443f-812f-a4932d5c595f
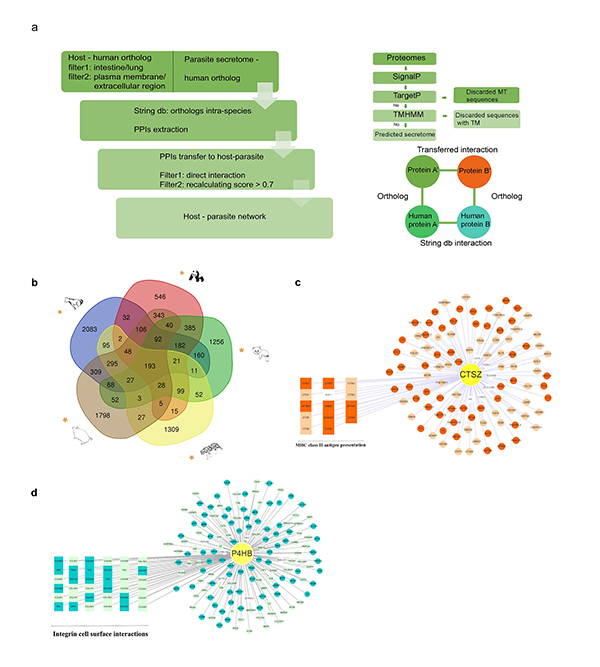
Figure 1. Analyses of genome-level protein interactions between five non-model mammals and their parasitic roundworms revealed that the immunoregulatory proteins CTSZ and P4HB played a central role in the protein interaction network.
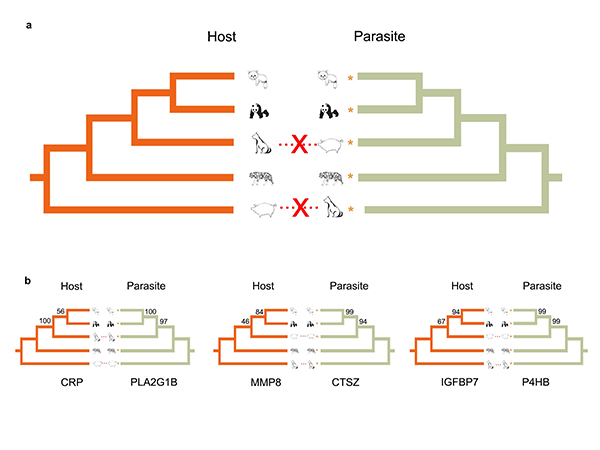
Figure 2. Phylogenetic relationships between mammalian hosts and their parasitic roundworms at the species-level (a) and gene-level (b), reflecting the gene-level coevolution between hosts and roundworms.
(Contact: Fuwen Wei, weifw@ioz.ac.cn )


