Organelles, which are basic components of cells, are formed by the separation of membrane (membrane-bound organelles) or via liquid-liquid phase separation (membraneless organelles). The membraneless feature allows membraneless organelles to easily communicate with cell matrix or other organelles, and even form or collapse in the response of organisms to stresses, modulating the cell resistance. However, little is known about the dynamic mechanism of membraneless organelles response to stress. The massive replications of viruses cause severe cellular stresses to host cells. The viral RNAs and proteins are thus targeted by host membraneless organelles. In the process of long-term adaptation, viruses change the accumulation of resistance-related membranous organelles in different ways to inhibit the host resistance system and facilitate the infections and proliferations of viruses. Therefore, virus infection provides a convenient system for studying the dynamic balance mechanism of membraneless organelles.
Now, researchers from the Institute of Zoology of the Chinese Academy of Sciences (CAS) have unraveled the dynamic of Dicing bodies (D-bodies), an important membraneless organelle that processes microRNA precursors into microRNAs, under normal conditions and in response to virus infection. The study was published in Science Advances.
The researchers found three RNA helicase proteins, RNA helicase 6 (RH6), RH8, and RH12, associate with the core components of D-bodies. The knockdown of these three genes results in a significant decrease, even disappearance, of D-bodies. These helicases interact with and facilitate the phase separation of SE, a core component of D-body, which subsequently promote the formation of D-bodies. Therefore, as novel components of D-body, RH6, RH8, and RH12 modulate D-bodies homeostasis through liquid-liquid phase separation (LLPS).
The researchers further studied the dynamic mechanism of membraneless organelles upon the infection of viruses. They found that the number of D-body in cells significantly decreases after viral infection. Meanwhile, after virus infection, the accumulations of RH6, RH8, and RH12 in the nucleus are significantly reduced, while the total amounts of these helicases remain unchanged, indicating viral infection alters the localizations of these RNA helicases and reduces their accumulation in the nucleus, resulting in the depolymerization of D-body. The researchers then found that these proteins are enriched at the viral replication sites. These helicases interact with viral protein VPg at viral replication sites and promote the phase separation of VPg to form a novel membraneless organelle, viral protein-containing bodies, and subsequently facilitate viral proliferation.
In this study, the researchers used D-body as a model to explore the mechanism of the formation of membraneless organelles and the dynamic of membraneless organelles in response to virus stress. These results will provide a theoretical basis for the development of new antiviral targets and the reduction of virus damage to crops.
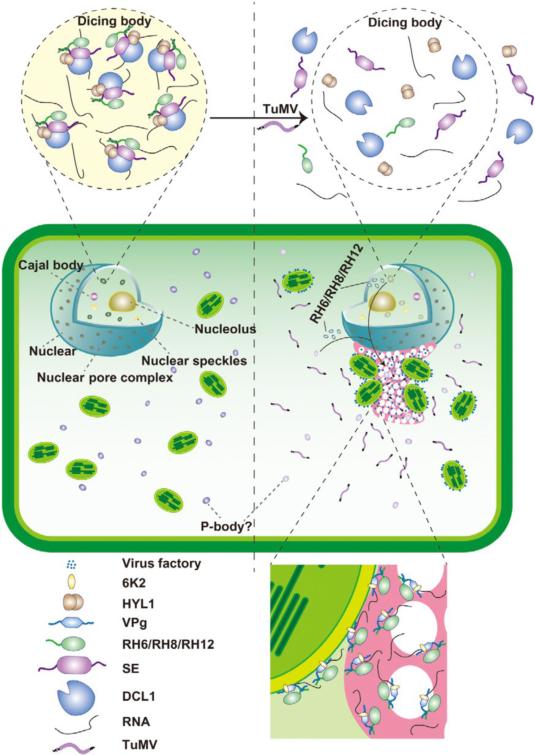
DEAD-box helicases drive the formation and virus-induced disassembly of dicing bodies
Contact: Xiaoming Zhang, zhangxm@ioz.ac.cn
Link: https://advances.sciencemag.org/content/7/18/eabc6266

