The high complexity and diversity of the eukaryotic transcriptome poses significant challenges for the efficient detection of specific transcripts. Conventional targeted RNA-seq methods often require labor-intensive pre-sequencing enrichment steps, which can compromise comprehensive transcriptome profiling and limit their broader applications.
Recently, Prof. ZHAO Fangqing’s team from the Institute of Zoology, Chinese Academy of Sciences, has developed a novel targeted RNA sequencing approach called PROgrammable Full-length Isoform Transcriptome sequencing (PROFIT-seq). This strategy effectively captures a broad spectrum of transcriptome samples, providing effective enrichment of target transcripts while maintaining unbiased quantification of the entire transcriptome.
PROFIT-seq introduces a programmable transcriptome sequencing strategy that leverages an AI-driven adaptive sequencing algorithm. Unlike conventional methods that rely on complex experimental enrichment, PROFIT-seq requires only the sequence information of the targets, enabling real-time, flexible, and programmable enrichment of different transcript types. It also employs a rolling circle amplification technique to generate full-length cDNA read-through products that facilitate correction of nanopore sequencing errors, resulting in high-fidelity consensus transcript sequences.
In addition, PROFIT-seq employs combinatorial reverse transcription to capture polyadenylated, non-polyadenylated, and circular RNAs. It also integrates an EM-based model to achieve unbiased transcriptome quantification using both fully sequenced targets and partially rejected sequences, providing efficient and accurate single-molecule characterization of any target transcript.
The research team further demonstrated the applicability of PROFIT-seq across various scenarios. In clinical samples, PROFIT-seq significantly increased the detection throughput of target pathogen transcripts and reduced the time required to identify key pathogen mutations through real-time enrichment of pneumonia-associated pathogens in sputum samples. In addition, PROFIT-seq enabled simultaneous detection of colorectal cancer-associated microbiota and host immune repertoire sequences, revealing intricate interactions between the host immune system and gut microbiota during the malignant transformation of colorectal polyps.
Overall, PROFIT-seq offers a powerful tool for accurate and efficient sequencing of target transcripts while preserving total transcriptome quantification, with broad potential for clinical diagnostics and targeted transcript enrichment scenarios.
This study entitled “Real-time and programmable transcriptome sequencing with PROFIT-seq” was published in Nature Cell Biology.
![]()
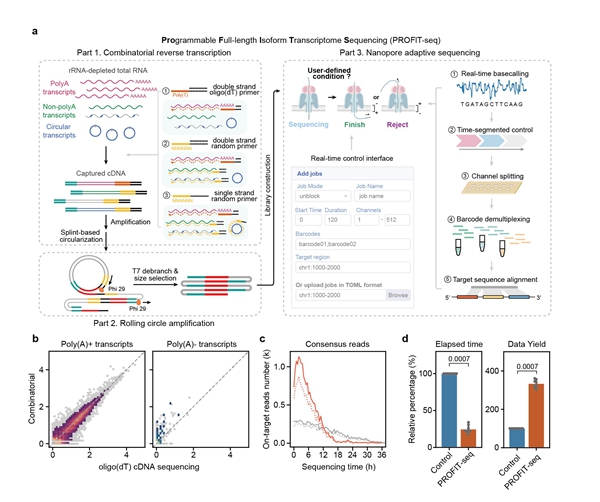
Figure: Programmable transcriptome sequencing enables highly efficient characterization of diverse target transcripts.
Article link: https://www.nature.com/articles/s41556-024-01537-1

