In an innovative study that could reshape our understanding of aging, a team of Chinese scientists from the Chinese Academy of Sciences and BGI Research has uncovered the intricate mechanisms by which immunoglobulins influence the aging process. This research, published in the prestigious journal Cell, not only charts a high-precision map of aging across various organs but also reveals the dual-edged sword of immunoglobulins in systemic aging.
The researchers constructed a detailed spatial transcriptomic landscape, dubbed Gerontological Geography (GG), which exposes the common threads of tissue structural disorder and loss of cellular identity as hallmarks of aging. This landscape is a significant step forward, pinpointing the epicenters of aging within multiple organs and uncovering the accumulation of immunoglobulins as a key aging characteristic and driver.
The quest for systemic biomarkers and key drivers of aging has been a long-standing puzzle in the field of gerontology. This study, a collaborative effort between Guang-Hui Liu's team from the Institute of Zoology, CAS, Ying Gu's team from BGI Research, Weiqi Zhang's team from the Beijing Institute of Genomics, CAS, and Jing Qu's team from the Institute of Zoology, CAS, has provided compelling answers.
By meticulously analyzing millions of spatial spots across nine organs in male mice, the team created high-precision spatial transcriptomic maps. These maps detailed the spatial distribution of over 70 cell types, offering a vivid picture of aging's spatial characteristics. Using a novel method of organizational structure entropy (OSE) analysis, they discovered that increased spatial structural disorder and loss of cellular identity are universal signs of systemic aging, suggesting that spatial structural damage may be a primary cause of organ functional decline during aging.
The team also identified senescence-sensitive spots (SSS), which are structural regions in different tissues more susceptible to aging's effects. They found that areas closer to SSS exhibit higher tissue structural entropy and greater loss of cellular identity, indicating that SSS could be the nucleus of organ aging. Notably, in immune organs, plasma cells, which are responsible for antibody synthesis, and cells with specific structures and functions, are the main components of the SSS microenvironment. The expression levels of immunoglobulin-related genes in these cells increase around SSS.
The study further discovered that immunoglobulin G (IgG) accumulates in multiple tissues and organs during aging in humans and mice, suggesting that IgG levels could serve as a new biomarker for aging. Moreover, IgG was found to directly induce aging in human and mouse macrophages and microglia, releasing inflammatory factors. Intriguingly, injecting IgG into young mice induced aging in multiple tissues and organs, demonstrating its potent aging effects.
In a promising development, the team developed an intervention strategy using antisense oligonucleotides (ASO) to reduce IgG content in mouse tissues, thereby delaying the aging of multiple organs.
This study is the first to map the spatial transcriptome of pan-organ aging in mammals, revealing tissue structural disorder and loss of cellular identity as key aging hallmarks and precisely locating the core regions and microenvironmental characteristics of aging sensitivity. The Immunoglobulin-associated Senescence Phenotype (IASP) proposed by the study expands the frontiers of aging science and opens new avenues for delaying aging and preventing related diseases.
The co-corresponding authors of the paper are Guang-Hui Liu from the Institute of Zoology, CAS, Ying Gu from BGI Research, Weiqi Zhang from the Beijing Institute of Genomics, CAS, and Jing Qu from the Institute of Zoology, CAS. The co-first authors include Shuai Ma, Zhejun Ji, Bin Zhang, Lingling Geng, Yusheng Cai, Chao Nie, Jiaming Li, Yuesheng Zuo, Yuzhe Sun, and Gang Xu.
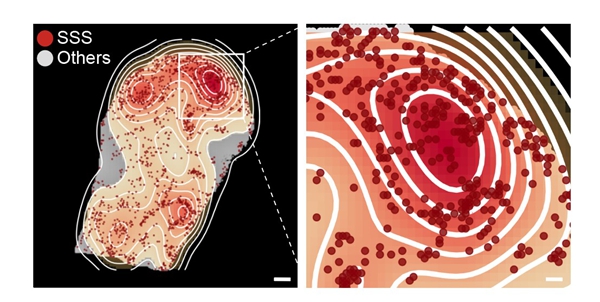
Figure 1. Spatial transcriptomic profiling reveals senescence-sensitive spots in the spleens of aged mice(Image by LIU Guanghui's lab)
Article link: https://doi.org/10.1016/j.cell.2024.10.019

